TO3-3PEG-Biotin Fluorophore
Specifications
| SKU | G959 | ||
| Name | TO3-3PEG-Biotin Fluorophore | ||
| Unit | 0.5 mg/ml (500 µl) | ||
| Description |
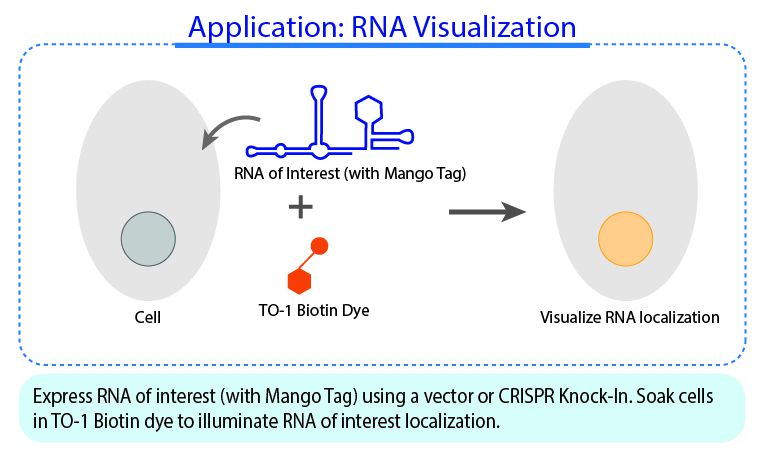
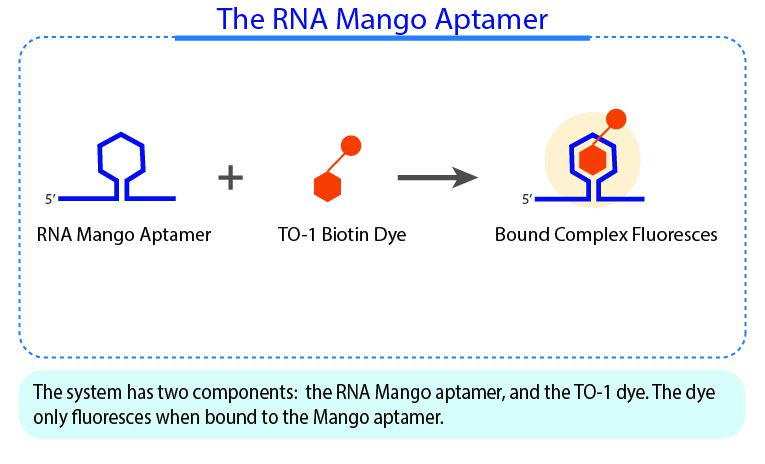
RNA Mango technology is based on the specific binding of the RNA Mango Aptamer and a Thizole Orange (TO) bi-functional dye. The main features of this technology is the tight binding between the dye and aptamer (KD ≈ 3nM) , and the strong ~1000X enhancement of the dye’s fluorescence when bound to the Mango apatmer (Fluorescent enhancement FE=1,100).The TO dye has a number of other desirable properties including:
Learn more about the RNA Mango technology here.
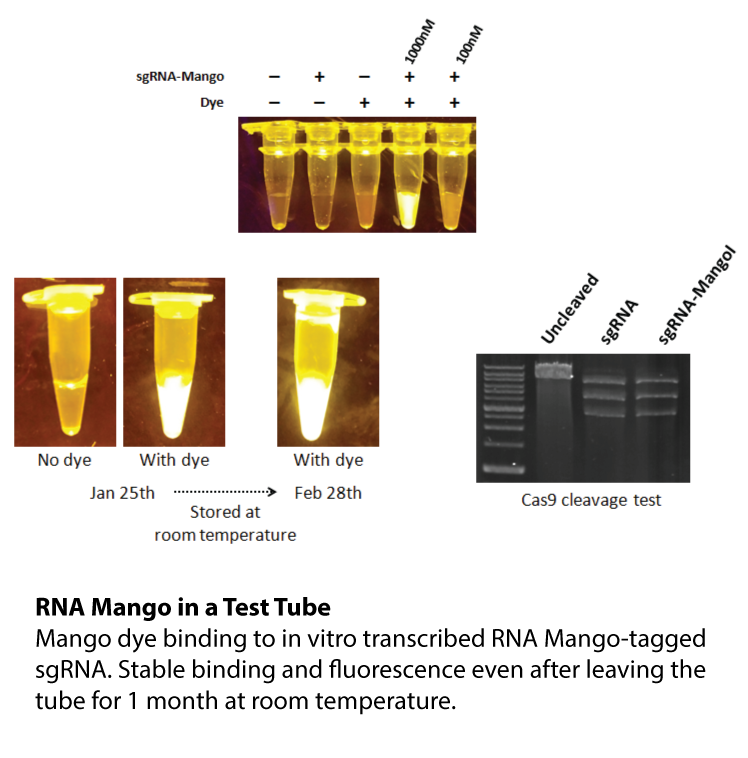
Watch RNA Mango in Action Transcription reaction were carried out in 300 µL volumes using T7 RNA polymerase (400 U, 50U/µL, applied biological materials), 0.5 µM TO1-3PEG-Biotin (applied biological materials), in 8 mM GTP, 5 mM CTP and ATP, 2 mM UTP, 40 mM TRIS buffer pH 7.9, 2.5 mM spermidine, 26 mM MgCl2, 20 mM KCl, Pyrophosphatase (0.5 U, 0.1 U/µL, ThermoFisher Scientific), and 0.01% Triton X-100. To each sample, either water (Negative), 0.33 µM DNA template (Mango Transcription), or 500 nM final Mango III A10U RNA (Positive) was added. Samples were visualized in a blue light box, movie is played back at 30X speed.
|
||
| Storage Condition |
Stored at -20°C and protect from light. |
FAQs
| Can I use water with my fluorophore dyes? | |
|
We strongly recommend to use DMF, DMSO, 10% Acetonitrile, or MeOH-CH₂ Cl₂ for stability. Do no store in water as it may break down the dye.
|
| Which aptamers should I use for each RNA Mango fluorophore? | |
|
Use our RNA Mango Aptamer Systems chart to help you select the appropriate aptamer and fluorophore combination for your research needs.
|
| Any tips for using RNA Mango aptamers in cellular transcripts? | |
|
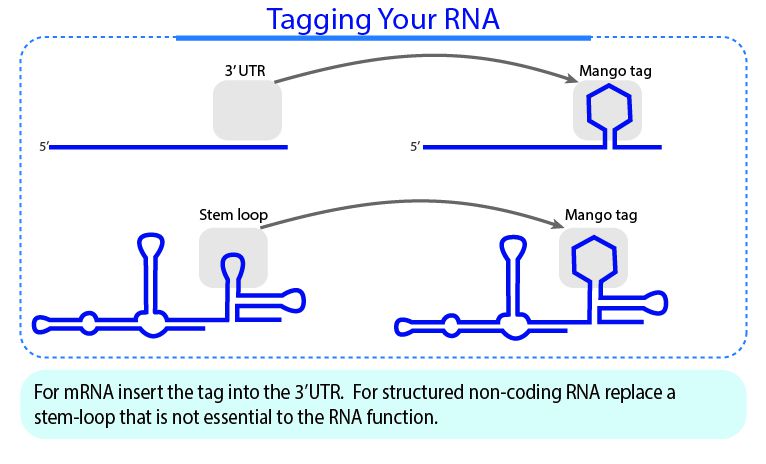
We have an aptamer insertion guideline for cellular transcript, which can be found here.
|
References
- Autour, A., C Y Jeng, S., D Cawte, A., Abdolahzadeh, A., Galli, A., Panchapakesan, S. S. S., Rueda, D., Ryckelynck, M., & Unrau, P. J. (2018). Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nature communications, 9(1), 656. https://doi.org/10.1038/s41467-018-02993-8
- Trachman, R. J., & Ferré-D'Amaré, A. R. (2019). Tracking RNA with light: selection, structure, and design of fluorescence turn-on RNA aptamers. Quarterly reviews of biophysics, 52, e8. https://doi.org/10.1017/S0033583519000064
- Trachman, R. J., 3rd, Autour, A., Jeng, S. C. Y., Abdolahzadeh, A., Andreoni, A., Cojocaru, R., Garipov, R., Dolgosheina, E. V., Knutson, J. R., Ryckelynck, M., Unrau, P. J., & Ferré-D'Amaré, A. R. (2019). Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nature chemical biology, 15(5), 472–479. https://doi.org/10.1038/s41589-019-0267-9
- Kong, K. Y. S., Jeng, S. C. Y., Rayyan, B., & Unrau, P. J. (2021). RNA Peach and Mango: Orthogonal two-color fluorogenic aptamers distinguish nearly identical ligands. RNA (New York, N.Y.), 27(5), 604–615. Advance online publication. https://doi.org/10.1261/rna.078493.120
- Cawte, A. D., Unrau, P. J., & Rueda, D. S. (2020). Live cell imaging of single RNA molecules with fluorogenic Mango II arrays. Nature communications, 11(1), 1283. https://doi.org/10.1038/s41467-020-14932-7
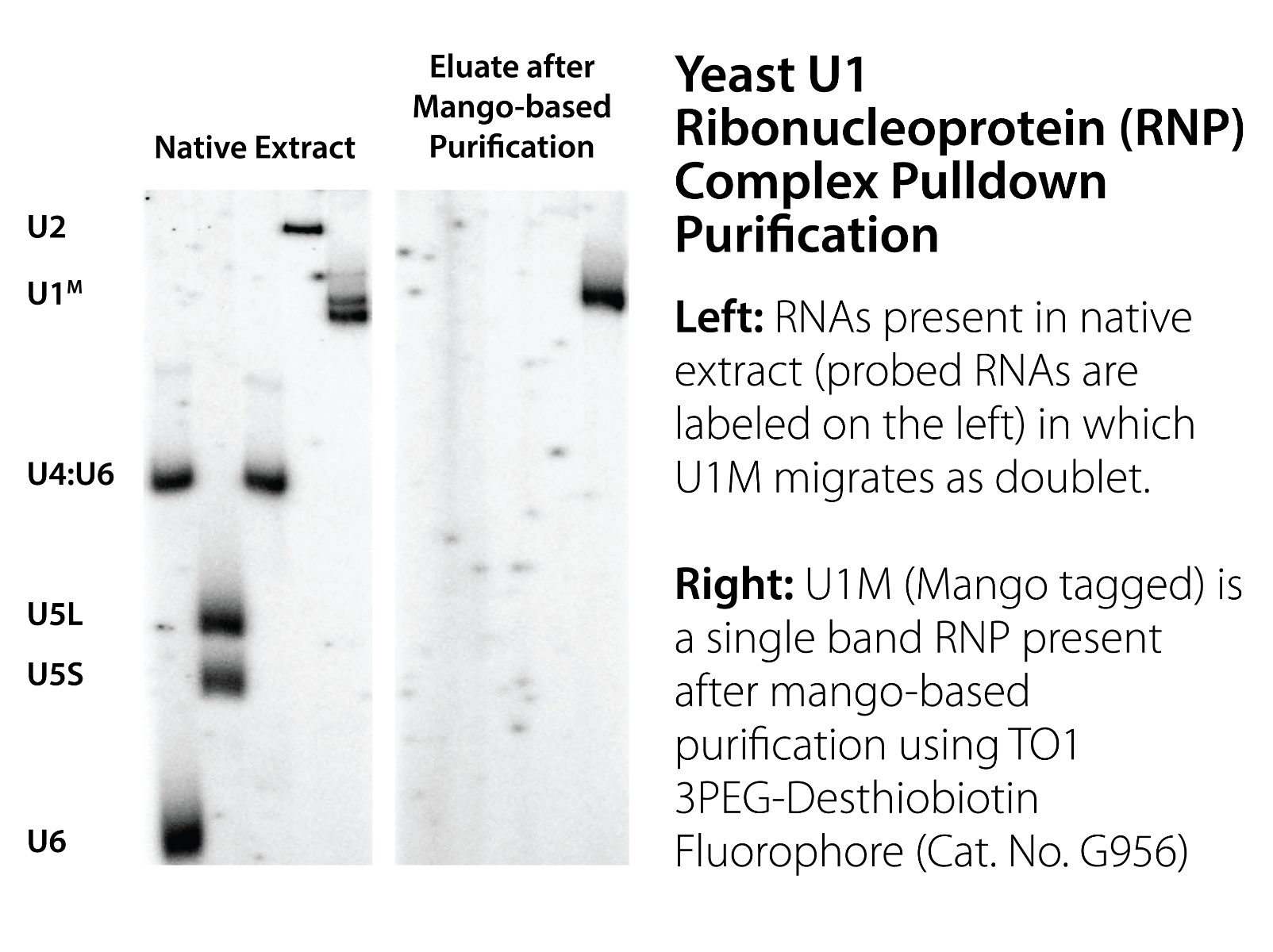
- Panchapakesan, S. S. S., Ferguson, M. L., Hayden, E. J., Chen, X., Hoskins, A. A., & Unrau, P. J. (2017). Ribonucleoprotein purification and characterization using RNA Mango. RNA (New York, N.Y.), 23(10), 1592–1599. https://doi.org/10.1261/rna.062166.117
- Mitra, J., & Ha, T. (2019). Nanomechanics and co-transcriptional folding of Spinach and Mango. Nature communications, 10(1), 4318. https://doi.org/10.1038/s41467-019-12299-y
- Shi, J., Gao, X., Tian, T., Yu, Z., Gao, B., Wen, A., You, L., Chang, S., Zhang, X., Zhang, Y., & Feng, Y. (2019). Structural basis of Q-dependent transcription antitermination. Nature communications, 10(1), 2925. https://doi.org/10.1038/s41467-019-10958-8
- Fang, C., Philips, S. J., Wu, X., Chen, K., Shi, J., Shen, L., Xu, J., Feng, Y., O'Halloran, T. V., & Zhang, Y. (2021). CueR activates transcription through a DNA distortion mechanism. Nature chemical biology, 17(1), 57–64. https://doi.org/10.1038/s41589-020-00653-x
Controls and Related Product: