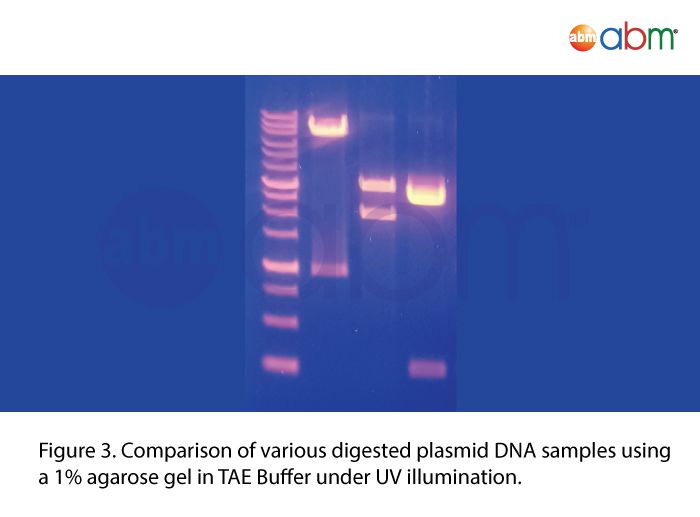
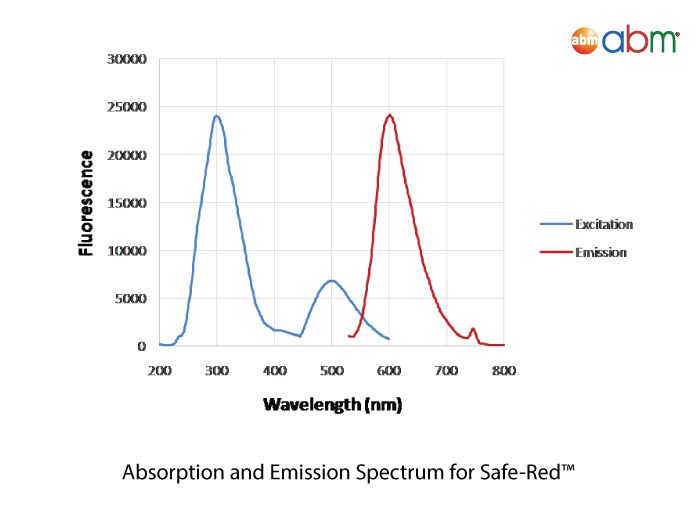
Safe-Red™
Specifications
| SKU | G108-R |
| Name | Safe-Red™ |
| Unit | 1.0 ml |
| Stain Color | Red |
| LED Viewer Compatibility | Yes |
| SafeView Series | Loading Dye |
| Description |
abm’s Safe-Red™ is a safe, improved formulation of nucleic acid stain for agarose gel electrophoresis. Safe-Red™ is offered in a convenient, one-step 6X loading dye format that allows for the elimination of toxic Ethidium Bromide and its associated contamination risks with glassware, gel apparatus and environment. Features:
|
| Applications |
Safe Detection of dsDNA, ssDNA and RNA in agarose gels. |
| Concentration | 6X |
| Storage Condition |
Store at 18-25°C. |
| Shipping Conditions |
Shipped on blue ice packs. |
| Note |
All SafeView DNA Stains are ISO-13485 certified. Dispose of SafeView DNA Stains as you would any other non-carcinogenic fluorescent dye (eg. Acridine orange; Propidium iodide). |
FAQs
| Can SafeView Classic be a replacement for ethidium bromide? Can I do gel extraction with it? | |
|
SafeView Classic can be used as a complete replacement for your ethidium bromide workflow. Yes, SafeView Classic can be used during gel extraction - in house testing demonstrated that gel extraction of DNA fragments using a SafeView Classic gel resulted in no decrease in ligation efficiency vs ethidium bromide. |
| How does SafeView work and why is it not carcinogenic? | |
|
abm's SafeView products contain fluorescent compounds that have a strong affinity to DNA and RNA nucleic acids. Once bound to nucleic acids, the compound fluoresces under specific wavelength of light which can then be visualized using a standard UV/Blue light imager. There may be some unknown effects of SafeView products that have not been documented in literature but these would also apply to popular SYBRSafe as well; however, SafeView products are not as carcinogenic as ethidium bromide. |
| How do I use SafeView products? | |
|
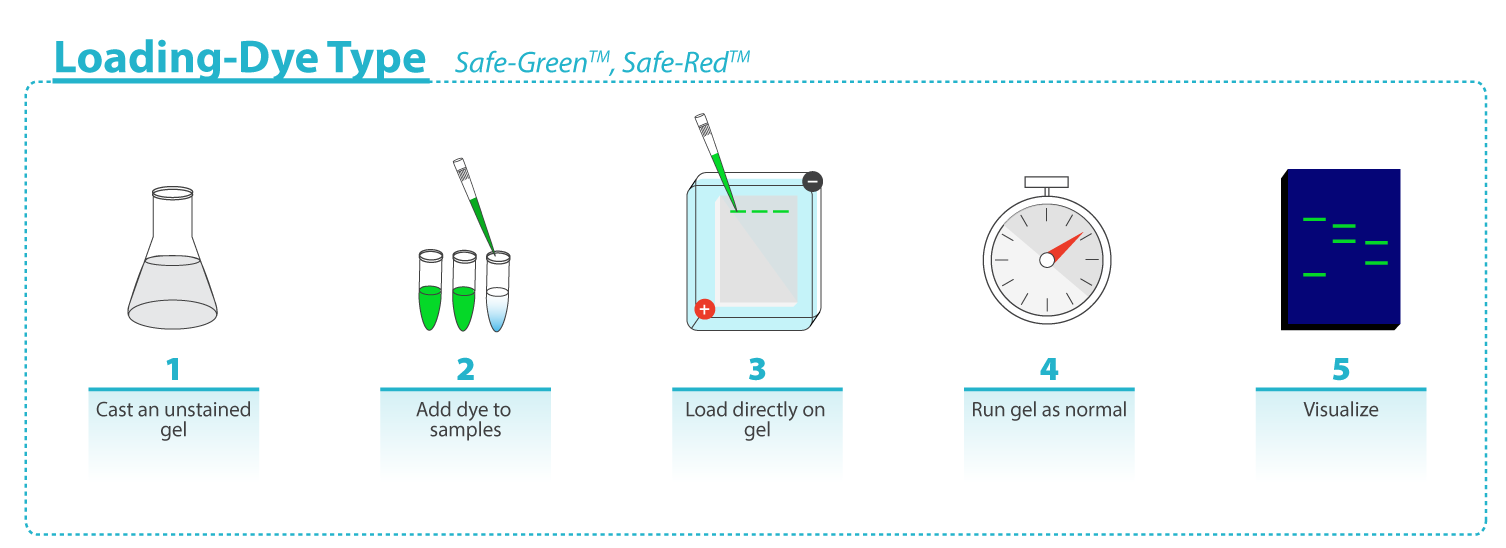
Safe-Red and Safe-Green are in a 6X loading dye format: mix samples and DNA marker with Safe-Red or Safe-Green at a 1:5 (dye : sample) dilution ratio, and load onto an unstained gel. SafeView Classic and Safe-Red Gel are in a 10,000X format: for pre-cast, add 10µl SafeView Classic or Safe-Red Gel per 100 ml molten agarose, mix gently and cast the gel; for post-stain, prepare and run an unstained agarose gel, next submerge the gel in post-staining solution of 30 μl SafeView Classic or Safe-Red Gel per 100 ml 1X TAE or 1X TBE buffer for 30min in the dark with light agitation, image accordingly. |
| Can the SafeView products differentiate double stranded vs single stranded nucleic acids? | |
|
No, our SafeView products will bind to both double-stranded and single-stranded nucleic acids, albeit with lesser efficiency for single-stranded nucleic acid species.
|
| At what temperature do I store the SafeView products? | |
|
SafeView Classic, Safe-Red and Safe-Red Gel should be stored at 18-25°C. Safe-Green should be stored at 4°C. |
| Do I need a special filter for photography of DNA gels stained with SafeView products? | |
|
All SafeView products are compatible with UV or Blue Light imagers. However, Safe-Green and SafeView Classic are more optimally visualized under Blue Light, making these stains safer for the user and their samples.
|
| How long does SafeView Classic stain last in a gel? | |
|
Our in-house testing has shown that SafeView Classic stained gels (>10ng DNA loaded per lane) can still be effectively visualized up to 1 week later with only a slight decrease in brightness. Gels should be stored properly to maintain a good signal, sealed in a plastic bag with wet paper towel loosely wrapped around the gel, away from light. Similarly, a pre-cast unused SafeView Classic gel can be stored and used up to 1 week after being prepared provided they are stored in ideal conditions (sealed in plastic bag, away from light).
|
| What is the sensitivity of the SafeView products? | |
|
Safe-Green: 0.2-0.6 ng DNA per band Safe-Red: 0.6-1.0 ng DNA per band Safe-Red Gel: 0.6-1.0 ng DNA per band SafeView Classic: 1.0-2.0 ng DNA per band |
| Can SafeView products be used to post-stain gels? | |
|
Yes, SafeView Classic and Safe-Red Gel can both be used for either pre-cast or post-stain gels.
|
| Why is the EtBr signal stronger in the pictures when I compare SafeView with EtBr? | |
|
Most gel imaging systems have been optimized for EtBr so this is a likely reason why the EtBr signal may be stronger in the pictures.
|
| Will I need an additional loading buffer for my samples? | |
|
If using Safe-Green or Safe-Red, you will not need to add any additional loading dyes or buffers. If using SafeView Classic or Safe-Red Gel, you will need to add an appropriate DNA loading dye to your samples in order for samples to properly settle and stay in the wells during electrophoresis. |
| We see migrations and band shifting of our fragments when using Safe-Green. Are there any recommendations that you can give us to minimize this band shifting? | |
|
Band shifting to a certain degree is unavoidable for any nucleic acid stain as most of these fluorescent compounds are large, positively charged molecules. Loading dye format nucleic acid stains, while convenient, may also suffer from more prominent band shifting due to the stain binding and migrating with the sample simultaneously during electrophoresis. Post-staining, while takes longer to complete, may be the optimal choice of nucleic acid stain strategy if band shifting issue is a priority. |
| I cannot see 100bp and 200bp bands on a 1% gel. What should I do? | |
|
It is often difficult to detect smaller 100bp and 200bp bands on a 1% gel especially if samples are of low concentration. We recommend visualizing smaller fragments on a higher concentration gel - ideally 2% agarose.
|
| Can I use SafeView products in polyacrylamide gels? | |
|
SafeView products are designed for use with agarose gels, and are not optimized for polyacrylamide gels.
|
| Which of the SafeView products will work with blue light / LED? | |
|
All of our SafeView products are compatible with UV and blue light.
|
| After the electrophoresis runs, is it okay to recover the sample stained with Safe-Red? Do I need to take special treatment to recover the sample DNA? I want to continue the cloning experiments with the sample after I check by electrophoresis. | |
|
Our Safe-Red™ (G108-R) will not affect the downstream cloning experiments, and thus no extra treatment is required to recover the stained sample DNA.
|
| What color is the tracking dye during electrophoresis? | |
|
Safe-Green and Safe-Red contain a blue tracking dye which will migrate at ~300bp after electrophoresis.
|
References
- Ibeagha-Awemu, EM et al. "Proteomics, genomics, and pathway analyses of Escherichia coli and Staphylococcus aureus infected milk whey reveal molecular pathways and networks involved in mastitis" J. Proteome Res. 9 (9):4604-4619 (2010). DOI: 10.1021/pr100336e. PubMed: 20704270. Application: PCR Products Viewing.
- Diaz-Balzac, CA et al. "Calbindin-D32k is localized to a subpopulation of neurons in the nervous system of the sea cucumber Holothuria glaberrima (Echinodermata)" PLoS ONE 7 (3):e32689 (2012). DOI: 10.1371/journal.pone.0032689. PubMed: 22412907. Application: PCR Products Viewing.
- Machida, RJ et al. "PCR primers for metazoan nuclear 18S and 28S ribosomal DNA sequences" PLoS ONE 7 (9):e46180 (2012). DOI: 10.1371/journal.pone.0046180. PubMed: 23049971. Application: PCR Products Viewing.
- Kim, NH et al. "Reactive oxygen species regulate context-dependent inhibition of NFAT5 target genes" Exp. Mol. Med. 45:e32 (2013). DOI: 10.1038/emm.2013.61.. PubMed: 23867654. Application: PCR Products Viewing.
- Goo, BG et al. "Bacillus thuringiensis: a specific gamma-cyclodextrin producer strain" Carbohydr. Res. 07-Dec:386 (2014). DOI: 10.1016/j.carres.2013.12.005. PubMed: 24456970. Application: PCR Products Viewing.
- Pyo, JS et al. "Activation of nuclear factor-κB contributes to growth and aggressiveness of papillary thyroid carcinoma" Pathol. Res. Pract. 209 (4):228-232 (2013). DOI: 10.1016/j.prp.2013.02.004. PubMed: 23528368. Application: PCR Products Viewing.
- Ish-Shalom, S et al. "Analysis of fungal gene expression by Real Time quantitative PCR" Methods Mol. Biol. 638:103-114 (2010). DOI: 10.1007/978-1-60761-611-5_7. PubMed: 20238263. Application: PCR Products Viewing.
- Lam, SW et al. "Rapid, specific and quantitative polymerase chain reaction (PCR) detection of pathogenic protozoa Entamoeba histolytica for drinking water supply" Water Science & Technology: Water Supply 11 (4):418-425 (2011). DOI: 10.2166/ws.2011.057. PubMed: 67657423. Application: PCR Products Viewing.
- Kim, SH et al. "The action of HIF-3a variants on HIF-2a–HIF-1ß heterodimer formation is directly probed in live cells" Exp Cell Res 336(2):329-37 (2015). DOI: 10.1016/j.yexcr.2015.06.017. PubMed: 26160453. Application: PCR Products Viewing.
- Vlasschaert, C et al. "Selection preserves Ubiquitin Specific Protease 4 alternative exon skipping in therian mammals" Sci. Rep. 6:20039 (2016). DOI: https://dx.doi.org/10.1038%2Fsrep20039.
- McCoy, P.. "Hormonally induced defects of DNA damage repair genes: an oncogenic driver of prostate cancer (Doctoral dissertation)" : (2018).
- Oguis, G. K.. "Clitoria ternatea (butterfly pea) cyclotides: insights on functional diversity, regulation and biotechnological applications" : (2019). DOI: 10.14264/uql.2019.610.
- Dickhout, . "U" S. Patent Application No. 0038597 A1 : (2019).
- Wang, B., Li, D., Cherkasova, V., Gerasymchuk, M., Narendran, A., Kovalchuk, I., & Kovalchuk, O. (2022). Cannabinol Inhibits Cellular Proliferation, Invasion, and Angiogenesis of Neuroblastoma via Novel miR-34a/tRiMetF31/PFKFB3 Axis. Cancers, 14(8), 1908. https://doi.org/10.3390/cancers14081908
- Wu, T., & Li, G. (2022). An Improved EMSA-based Method to Prioritize Candidate cis-REs for Further Functional Validation. BIO-PROTOCOL, 12(8). https://doi.org/10.21769/bioprotoc.4397
Controls and Related Product: