ATP2C2 AAV Vector (Human) (CMV)
Specifications
Print Custom Datasheet
Documents
Supporting Protocol
MSDS
FAQs
| What are the correct concentration units for each recombinant viral particle? | |
|
For lentiviruses and retroviruses, they are measured in CFU/ml (colony-forming units per millilitre). Transduction with lentiviruses and retroviruses can cause the formation of colonies, which can be quantified for concentration. For AAV the titer is measured as genome copies per mL (GC/mL). Adenoviruses are measured as PFU/ml (plaque-forming units per millilitre). Transduction with adenoviruses will kill packaging cells, forming plaques in the process for quantification. The concentration for all three types of viruses can also be classified as IU/ml (Infectious Units/ml). Ultimately, the units refers to the viral particles and different units reflect the different assays involved.
|
| What do I use to check if my cells were successfully immortalized by the SV40 agent? | |
|
We have an SV40 T antibody that can be used for the western blot analysis. The catalog number is G202.
Otherwise, a qPCR primer can be designed on the SV40 gene for qPCR analysis. The sequence can be found in the link below:
http://www.abmgood.com/pLenti%20SV40-Vector-Location-Map.html
|
| What are the primers to use for SV40 identification? | |
|
SV40 Forward Primer Sequence
5’ ACTGAGGGGCCTGAAATGA
SV40 Reverse Primer Sequence
5’ GACTCAGGGCATGAAACAGG
These are qPCR primers and the band size is 61 bp.
|
| What advantages / disadvantages exist between the Lenti-SV40, -SV40T, and SV40T+t vectors? | |
|
There are simply differences in the content of all vectors due to customer demand for variety. Lenti-SV40 will contain the whole SV40 gene, -SV40T, the large T Antigen only, and -SV40T&t the large and small T antigens only.
It is up to the end user to decide which vectors will best suit their project, however we have successfully used Lenti-SV40 (whole gene) in a wide range of immortalization projects.
|
| What is the accession number for the SV40? | |
|
The SV40 covers the entire genome and the accession number is J02400.1. You can use this information to design primers for conventional PCR as well.
|
| How long after transduction can the infection efficiency be observed? | |
|
You can observe transduction efficiency from 48 hours up to 5 days after infection.
|
| What are the primers to use for SV40T and SV40T tsA58 detection? | |
|
PCR primers:
SV40T Forward Primer Sequence
5’ AGCCTGTAGAACCAAACATT 3'
SV40T Reverse Primer Sequence
5’ CTGCTGACTCTCAACATTCT 3'
The two primers should amplify the region between 3677-4468bp, giving a 792bp fragment.
|
| What is the sequence of the SV40 large T antigen? | |
|
This information can be accessed on this page by clicking on "pLenti-SV40-T" under vector map. The Large T antigen is at position 5079-5927.
|
| How much DNA do I need to provide for Custom AAV without DNA Amplification? | |
|
You will need to provide purified plasmid DNA at a concentration of 0.5ug/ul or more.
50 ug DNA needed for Custom AAV without DNA amplification (10^9 GC/ml) 200 ug DNA needed for Custom AAV without DNA amplification (10^12 GC/ml) 300 ug DNA needed for Custom AAV without DNA amplification (10^13 GC/ml) |
| What's the difference between GC/ml and vg/ml? | |
|
Both are used interchangeably and it is a qPCR based titer method.
|
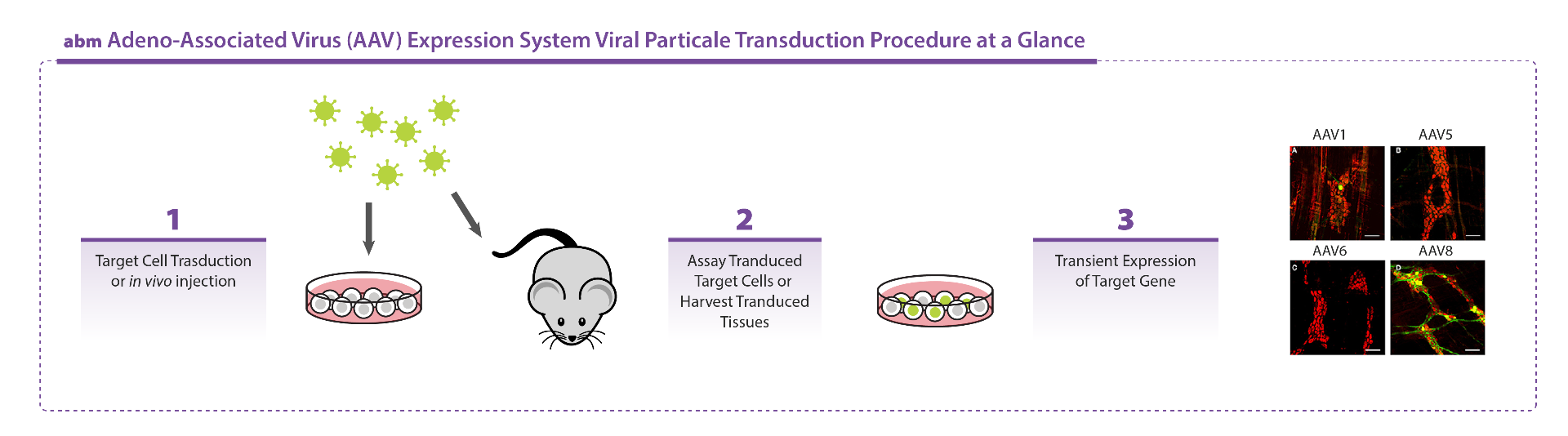
| Why do I not see GFP expression after infecting AAV in my cell line? | |
|
Serotype selection is an important parameter affecting transduction ability of AAV particles. Thus, determine which serotype works best for your cell line. You could refer to our technical sheet "AAV - General Guideline to Serotype Selection". eg. Serotype 5 has limited transduction ability on most cell types.
|
| Is your AAV preparation in-vivo grade? | |
|
Our high titer AAV preps undergo extensive purification steps leading to high quality viral particles ready to inject for your in-vivo models. For reference, see our in-vivo infectivity data as tested by an independent lab {https://www.abmgood.com/AAV-Adeno-Associated-Virus.html}
|
| For G221 and LV620, what does the 'V12' in RasV12 mean? | |
|
The V12 means that amino acid # 12 is mutated from a Valine to a Glycine. Other than that, the sequence matches the coding region of HRAS perfectly (NM_005343).
|
| Where is the SV40T tsA58 gene sequence? | |
|
The SV40T tsA58 gene is located between 3138-5264bp, with the Alanine-to-Valine mutation at amino acid 438.
|
| What are the QC methods for testing AAV? | |
|
We provide qPCR based titer as primary method of determining successful packaging. If your virus has GFP or RFP reporter, we also perform virus infectivity testing in HEK293T cell line and provide you image in CoA.Since infectivity is serotype dependent, and HEK293T cells are not infected well with Serotype 4,5 and 6, we are unable to confirm infection for these serotypes in our standard evaluation cell line. For such cases, titer as determined by qPCR is considered final parameter to determine successful packaging of virus.
|
| Can I work with different serotypes of AAV virus in the same equipment to keep the infected cells? | |
|
There is no problem with using different serotypes in the same equipment, as long as the handler takes the basic precautions to avoid cross-contamination.
|
References
- Iacono, L. L., Lelpo, D., Accoto, A., Di Segni, M., Babicola, L., D’Addario, S. L., ... & Andolina, D. "MicroRNA-34a Regulates the Depression-like Behavior in Mice by Modulating the Expression of Target Genes in the Dorsal Raphè" Molecular neurobiology 1-14: (2019).
Other ATP2C2 Products
ATP2C2
Human
ATP2C2
Mouse
ATP2C2
Rat
ATP2C2
Lentiviral
Vector
ATP2C2
AAV
Vector
ATP2C2
Adenovirus
Vector
ATP2C2
Protein
Vector
ATP2C2
ORF
Vector
ATP2C2
siRNA
Vector
ATP2C2
miRNA
Vector
ATP2C2
3'UTR
Vector
ATP2C2
CRISPR Knockout
Vector
ATP2C2
CRISPR Activation
Vector
ATP2C2
Control Vectors & Viruses
Vector
ATP2C2
circRNA
Vector
ATP2C2
Internal
Vector